Molecular Docking Screening and Pharmacokinetic Studies of Some Boron-Pleuromutilin Analogues against Possible Targets of Wolbachia pipientis
Main Article Content
Abstract
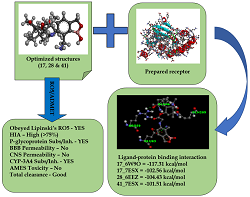
Lymphatic filariasis and onchocerciasis are two common filarial diseases caused by a group of parasitic nematodes called filarial worms, which co-habit with the bacteria organism Wolbachia. One good treatment approach seeks Wolbachia as a drug target. Here, a computer-aided molecular docking screening was conducted on a series of 52 pleuromutilin analogs against four Wolbachia enzymes: α-DsbA1 (PDB: 3F4R), α-DsbA2 (6EEZ), OTU deubiquitinase (6W9O), and cytoplasmic incompatibility factor CidA (7ESX) to find a more potent drug candidate(s) for the treatment of filarial diseases. The docking investigation was performed using the iGEMDOCK tool, while NAMD was utilized for the Molecular Dynamic (MD) simulation. The results of the virtual screening identified four ligand-protein interaction pairs with the highest binding affinities in the order: 17_6W9O (-117.31 kcal/mol) > 28_6EEZ (-104.43 kcal/mol) > 17_7ESX (-102.56 kcal/mol) > 41_7ESX (-101.51 kcal/mol), greater than that of the reference drug doxycycline_7ESX (-92.15 kcal/mol). These molecules (17, 28, and 41) showed excellent binding interactions, making very close contact with the receptors’ amino acid residues. They also showed better pharmacokinetic properties than doxycycline because they showed high intestinal absorption, were orally bioavailable and showed no AMES toxicity. Also, the stability of 17_6W9O interactions was confirmed by the MD simulation. Therefore, the selected molecules could be developed as potential drug candidates for treating filarial diseases.
Downloads
Article Details

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Authors continue to retain the copyright to the article if the article is published in the Journal of Molecular Docking. They will also retain the publishing rights to the article without any restrictions.
Authors who publish with this journal agree to the following terms:
- Any article on the copyright is retained by the author(s).
- The author grants the journal, right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share work with an acknowledgment of the work authors and initial publications in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of published articles of work (eg, post-institutional repository) or publish it in a book, with acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their websites) prior to and during the submission process, as can lead to productive exchanges, as well as earlier and greater citation of published work.
- The article and any associated published material are distributed under the Creative Commons Attribution-ShareAlike 4.0 International License.
References
Mitra AK, Mawson AR. Neglected Tropical Diseases: Epidemiology and Global Burden. Trop Med Infect Dis. 2017;2(3):36. doi:10.3390/tropicalmed2030036
Bakowski MA, Shiroodi RK, Liu R, Olejniczak J, Yang B, Gagaring K, et al. Discovery of short-course antiwolbachial quinazolines for elimination of filarial worm infections. Sci Transl Med. 2019;11(491): eaav3523. doi:10.1126/scitranslmed.aav3523
Jacobs RT, Lunde CS, Freund YR, Hernandez V, Li X, Xia Y, et al. Boron-pleuromutilins as anti-wolbachia agents with potential for treatment of onchocerciasis and lymphatic filariasis. J Med Chem. 2019;62:2521−40. doi:10.1021/acs.jmedchem.8b01854
Carter DS, Jacobs RT, Freund Y, Berry P, Akama T, Easom EE, et al. Macrofilaricidal benzimidazole-benzoxaborole hybrids as an approach to the treatment of river blindness, part 2: ketone linked analogs. ACS Infect Dis. 2020;6(2):180-5. doi:10.1021/acsinfecdis.9b00397
Weil GJ, Bogus J, Christian M, Dubray C, Djuardi Y, Fischer PU, et al. The safety of double- and triple-drug community mass drug administration for lymphatic filariasis: A multicenter, open-label, cluster-randomized study. PLoS Med. 2019;16(6):e1002839. doi:10.1371/journal.pmed.1002839
Sashidhara KV, Rao KB, Kushwaha V, Modukuri RK, Verma R, Murthy PK. Synthesis and antifilarial activity of chalcone-thiazole derivatives against a human lymphatic filarial parasite, Brugia malayi. Eur J Med Chem. 2014;81:473-80. doi:10.1016/j.ejmech.2014.05.029
Slatko BE, Taylor MJ, Foster JM. The wolbachia endosymbiont as an anti-filarial nematode target. Symbiosis. 2010;51(1):55−65. doi:10.1007/s13199-010-0067-1
Bouchery T, Lefoulon E, Karadjian G, Nieguitsila A, Martin C. The symbiotic role of wolbachia in onchocercidae and its impact on filariasis. Clin Microbiol Infect. 2013;19(2):131–40. doi:10.1111/1469-0691.12069
Bakowski MA, McNamara CW. Advances in Antiwolbachial Drug Discovery for Treatment of Parasitic Filarial Worm Infections. Trop Med Infect Dis. 2019;4(3):108. doi:10.3390/tropicalmed4030108
Sulaiman WAW, Kamtchum-Tatuene J, Mohamed MH, Ramachandran V, Ching SM, Lim SMS, et al. Anti- Wolbachia therapy for onchocerciasis & lymphatic filariasis: Current perspectives. Indian J Med Res. 2019;149(6):706-14. doi:10.4103/ijmr.ijmr_454_17
Newman DJ, Cragg GM. 2.19 - Natural Products of Therapeutic Importance. Compr Nat Prod II. 2010;2:623-50. doi:10.1016/B978-008045382-8.00055-1
Dasenaki ME, Thomaidis, NS. Chapter 18 - Meat Safety: II Residues and Contaminants. Lawrie´s Meat Sci. 2017;553–83. doi:10.1016/B978-0-08-100694-8.00018-2
Brown P, Dawson MJ. Chapter Three - A Perspective on the Next Generation of Antibacterial Agents Derived by Manipulation of Natural Products. Prog Med Chem. 2015;54:135–84. doi:10.1016/bs.pmch.2014.10.001
Paukner S, Riedl R. Pleuromutilins: Potent Drugs for Resistant Bugs-Mode of Action and Resistance. Cold Spring Harb Perspect Med. 2017;7(1):a027110. doi:10.1101/cshperspect.a027110
Novak R, Shlaes DM. The pleuromutilin antibiotics: a new class for human use. Curr Opin Investig Drugs. 2010;11(2):182-91.
Adeniji SE, Arthur DE, Abdullahi M, Abdullahi A, Ugbe FA. Computer-aided modeling of triazole analogues, docking studies of the compounds on DNA Gyrase enzyme and design of new hypothetical compounds with efficient activities. J Biomol Struct Dyn. 2020;40(9):4004-20. doi:10.1080/07391102.2020.1852963
Ibrahim MT, Uzairu A, Shallangwa GA, Uba S. Lead identification of some anti-cancer agents with prominent activity against Non-Small Cell Lung Cancer (NSCLC) and structure-based design. Chem Afr. 2020;3:1023-44. doi:10.1007/s42250-020-00191-y
Lawal HA, Uzairu A, Uba S. QSAR, molecular docking studies, ligand-based design and pharmacokinetic analysis on Maternal Embryonic Leucine Zipper Kinase (MELK) inhibitors as potential anti-triple-negative breast cancer (MDA-MB-231cell line) drug compounds. Bull Natl Res Cent. 2021;45:90. doi:10.1186/s42269-021-00541-x
Ibrahim MT, Uzairu A, Uba S, Shallangwa GA. Design of more potent quinazoline derivatives as EGFRWT inhibitors for the treatment of NSCLC: A computational approach. Future J Pharm Sci. 2021;7:140. doi:10.1186/s43094-021-00279-3
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1-3):3–26. doi:10.1016/s0169-409x(00)00129-0
Edache EI, Uzairu A, Mamza PA, Shallangwa GA. Computational modeling and analysis of the theoretical structure of thiazolino 2 pyridone amide inhibitors for Yersinia pseudo-tuberculosis and Chlamydia trachomatis Infectivity. Bull Sci Res. 2022;4(1):14-39. doi:10.54392/bsr2212
Lu L, Hu H, Hou H, Wang B. An improved B3LYP method in the calculation of organic thermochemistry and reactivity. Comput Theor Chem. 2013;1015:64-71. doi:10.1016/j.comptc.2013.04.009
Li Z, Wan H, Shi Y, Ouyang P. Personal experience with four kinds of chemical structure drawing software: review on ChemDraw, ChemWindow, ISIS/Draw, and ChemSketch. J Chem Inf Comput Sci. 2004;44(5):1886-90. doi:10.1021/ci049794h
Ugbe FA, Shallangwa GA, Uzairu A, Abdulkadir I. Theoretical activity prediction, structure‑based design, molecular docking and pharmacokinetic studies of some maleimides against Leishmania donovani for the treatment of leishmaniasis. Bull Natl Res Cent. 2022;46:92. doi:10.1186/s42269-022-00779-z
Hsu K, Chen Y, Lin S, Yang J. iGEMDOCK: a graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinform. 2011;12(Suppl 1):S33. doi:10.1186/1471-2105-12-S1-S33
Ugbe FA, Shallangwa GA, Uzairu A, Abdulkadir I. Activity modeling, molecular docking and pharmacokinetic studies of some boron-pleuromutilins as anti-wolbachia agents with potential for treatment of filarial diseases. Chem Data Collect. 2021;36:100783. doi:10.1016/j.cdc.2021.100783
Du X, Li Y, Xia YL, Ai SM, Liang J, Sang P, et al. Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods. Int J Mol Sci. 2016;17(2):144. doi:10.3390/ijms17020144
Sun Y, Yang AW, Hung A, Lenon GB. Screening for a potential therapeutic agent from the herbal formula in the 4th edition of the Chinese national guidelines for the initial-stage management of COVID-19 via molecular docking. Evid Based Complement Alternat Med. 2020;2020:3219840. doi:10.1155/2020/3219840
Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. 2016;12(1):405-13. doi:10.1021/acs.jctc.5b00935
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781-802. doi:10.1002/jcc.20289
Edache EI, Uzairu A, Mamza PA, Shallangwa GA. A comparative QSAR analysis, 3D-QSAR, molecular docking and molecular design of iminoguanidine-based inhibitors of HemO: A rational approach to antibacterial drug design. J Drugs Pharm Sci. 2020;4(3):21-36. doi:10.31248/JDPS2020.036