Identification of Bioactive Molecules from Combretum micranthum as Potential Inhibitors of α-amylase through Computational Investigations
Main Article Content
Abstract
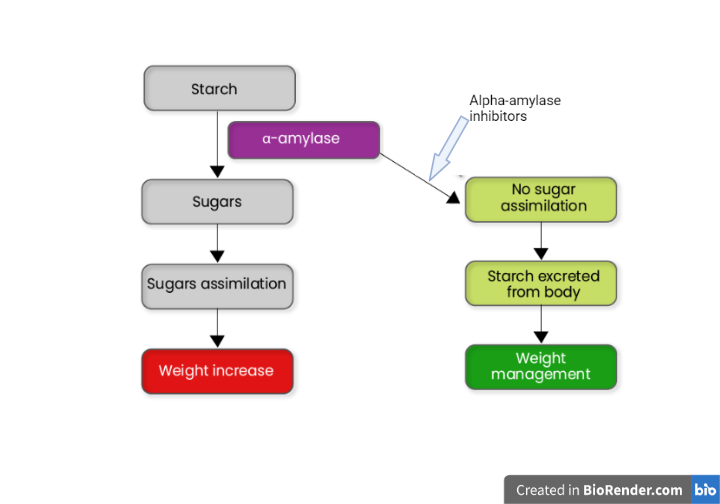
The rising prevalence of diabetes necessitates continued research into natural antidiabetic medicines that target a key biochemical enzyme involved. The α-amylase enzyme is involved in the digestion of starch, glycogen, and disaccharides in the gastrointestinal tract. Its essential roles and distinct properties make it an effective antidiabetic target. This work aimed to use in silico approaches to find possible α-amylase inhibitors from Combretum micranthum bioactive substances. On the Schrödinger Maestro 12.5, over 50 C. micranthum compounds were screened, followed by MM-GBSA and ADMET (absorption, distribution, metabolism, excretion, and toxicity) studies of the highest affinity compounds. The α-amylase binding affinities were higher for rutin trihydrate and myricetin-3-rutinoside (-12.162 kcal/mol and -10.935 kcal/mol, respectively). They reacted with amino acids that are required for the inhibition of α-amylase. As a result, these compounds have the structural characteristics, binding affinities, and molecular interactions necessary as α-amylase inhibitors and could be turned into antidiabetic medicines through lead optimization and experimental research.
Downloads
Article Details

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Authors continue to retain the copyright to the article if the article is published in the Journal of Molecular Docking. They will also retain the publishing rights to the article without any restrictions.
Authors who publish with this journal agree to the following terms:
- Any article on the copyright is retained by the author(s).
- The author grants the journal, right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share work with an acknowledgment of the work authors and initial publications in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of published articles of work (eg, post-institutional repository) or publish it in a book, with acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their websites) prior to and during the submission process, as can lead to productive exchanges, as well as earlier and greater citation of published work.
- The article and any associated published material are distributed under the Creative Commons Attribution-ShareAlike 4.0 International License.
References
To be published