De Novo Class of Momordicoside with Potent and Selective Tumor Cell Growth Inhibitory Activity as Pyruvate Kinase Muscle Isozyme 2 and Anti-apoptotic Myeloid Leukemia 1 Inhibitors
Main Article Content
Abstract
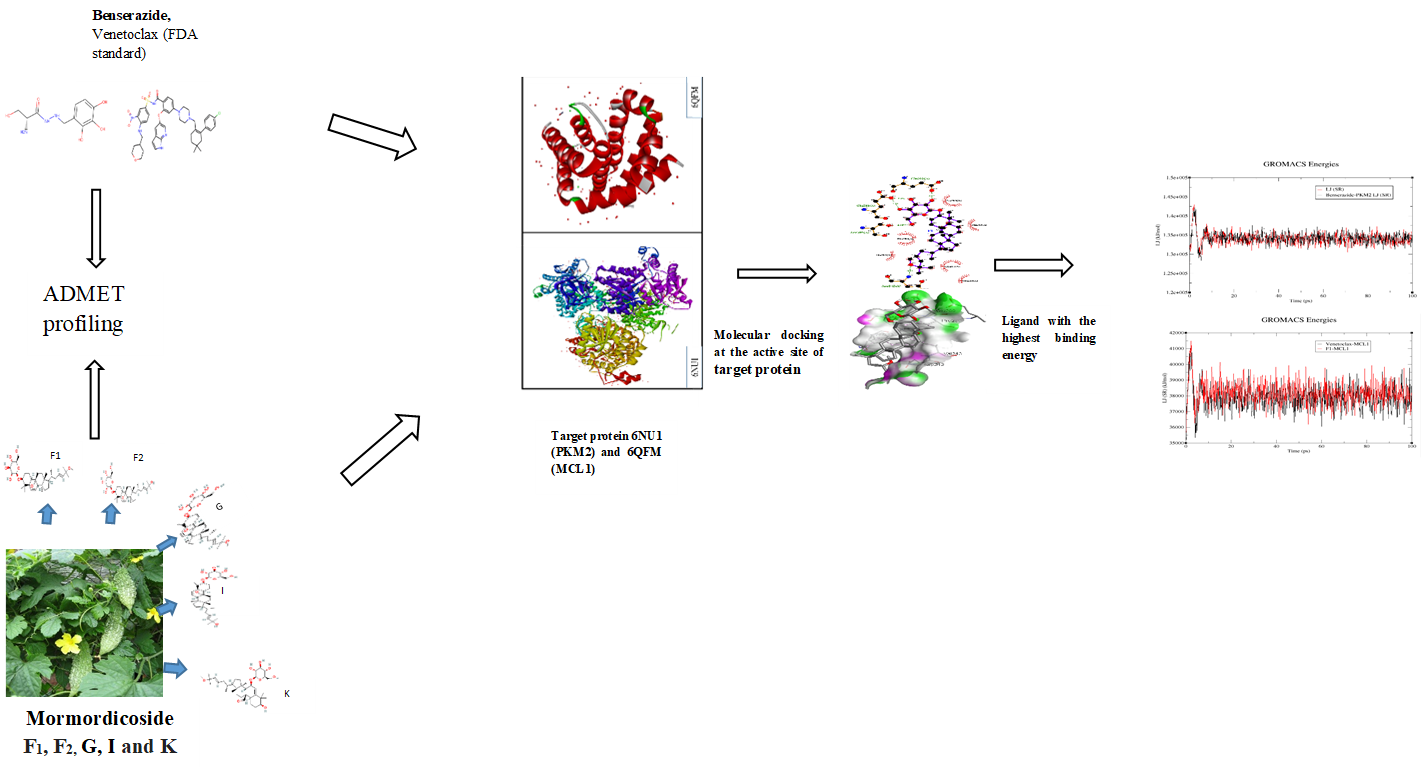
The difficulty in treating cancer resides in targeting abnormal proliferation while protecting normal proliferation, necessitating a thorough comprehension of the normal and malignant mechanisms that promote cell growth and proliferation. Targeting cell death signaling pathways such as glycolytic and mitochondrial apoptosis is the hallmark of many cancers, the aim of which this research is ready to evaluate. Atomistic molecular dynamics simulation of top hits after molecular docking and ADMET profiling of the ligands were performed for main protease-hit complexes. Docking scores of ligands used against PKM2 range from –9.36 to –12.1 kcal/mol, wherein momordicoside-F2 had the highest score (2.1 kcal/mol), performing better than the FDA-approved drug benserazide. Likewise, the scores ranged between –8.51 and –12.05 kcal/mol for Anti-apoptotic Myeloid Leukemia 1 (MCL-1), with momordicoside-F1 being the highest-ranked compound. The RMSD plots depicted stable trajectories with consistent and minor fluctuations implying that the protein (PKM2 and MCL1) backbone underwent minor structural perturbations. In addition, several significant peaks of increased fluctuations were also observed, indicating their increased interaction potential, implying that the ligands could adapt well in the protein's binding pocket. The SASA analysis results show that the ligands retained inside their shallow binding pocket. The phylogenetic tree obtained implies the likelihood of recurring results of the in silico profiling. Conclusively, this research unveils that Mormordicoside F1 shows good stability with MCL-1, likewise, momordicoside-F2 against PKM2. These hits can be a better re-purposing option.
Downloads
Article Details

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Authors continue to retain the copyright to the article if the article is published in the Journal of Molecular Docking. They will also retain the publishing rights to the article without any restrictions.
Authors who publish with this journal agree to the following terms:
- Any article on the copyright is retained by the author(s).
- The author grants the journal, right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share work with an acknowledgment of the work authors and initial publications in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of published articles of work (eg, post-institutional repository) or publish it in a book, with acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their websites) prior to and during the submission process, as can lead to productive exchanges, as well as earlier and greater citation of published work.
- The article and any associated published material are distributed under the Creative Commons Attribution-ShareAlike 4.0 International License.
References
To be published