Coronavirus-SARS-CoV-2: Biology and Problems in rRT-PCR Detection
Abstract
Coronavirus disease 2019 (COVID-19) first appeared in China in December 2019 and was declared a pandemic by the World Health Organization. COVID-19 is caused by Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2), a new virus previously unknown to humans. Here we look at what is known about this virus, the main method for detecting the presence of this virus in a person who is used as a golden standard, and the problems that could arise in this detection method. Understanding the biology of the virus and the strengths and weaknesses of the detection method are important for patient management and for overcoming the pandemic.
Full text article
References
Bustin, S.A. & Nolan, T. (2004). Pitfalls of Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction. Journal of Biomolecular Techniques, 15(3), 155-166.
Center for Disease Control and Prevention. (2020a). CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. https://www.fda.gov/media/134922/download
Center for Disease Control and Prevention. (2020b). Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens for COVID-19. https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html
Corman, V.M., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D.K.W., Bleicker, T., Brunink, S., Schneider, J., Schmidt, M.L., Mulders, D.G.J.C., Haagmans, B.L., van der Veer, B., van den Brink, S., Wijsman, L., Goderski, G., Romette, J.L., Ellis, J., Zambon, M., Peiris, Goossens, H., Reusken, C., Koopmans, M.P.G., & Drosten, C. (2020). Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance, 25(14), 20200409c. doi:10.2807/1560-7917.ES.2020.25.14.20200409c
Deepak, S.A., Kottapalli, K.R., Rakwal, R., Oros, G., Rangappa, K.S., Iwahashi, H., Masuo, Y., & Agrawal, G.K. (2007). Real-Time PCR: Revolutionizing Detection and Expression Analysis of Genes. Current Genomics, 8(4), 234–251. doi:10.2174/138920207781386960
Espy, M.J., Uhl, J.R., Sloan, L.M., Buckwalter, S.P., Jones, M.F., Vetter, E.A., Yao. J.D.C., Wengenack, N.L., Rosenblatt, J.E., Cockerill, F.R., & Smith, T.F. (2006). Real-Time PCR in Clinical Microbiology: Applications for Routine Laboratory Testing. Clinical Microbiology Reviews, 19(1), 165-256. doi:10.1128/CMR.19.1.165-256.2006
Forster, P., Forster, L., Renfrew, C., & Forster, M. (2020). Phylogenetic Network Analysis of SARS-CoV-2 Genomes. Proceedings of the National Academy of Science of the United States of America, 117(17), 9241-9243. doi:10.1073/pnas.2004999117
Garibyan, L., & Avashia, N. (2013). Research Techniques Made Simple: Polymerase Chain Reaction (PCR). Journal of Investigative Dermatology, 133(3), e6. doi:10.1038/jid.2013.1
Guan, W., Ni, Z., Hu, Y., Liang, W., Ou, C., He, J., Liu, L., Shan, H., Lei, C., Hui, D. S. C., Du, B., Li, L., Zeng, G., Yuen, K. Y., Chen, R., Tang, C., Wang, T., Chen, P., Xiang, J., & Zhong, N. (2020). Clinical Characteristics of Coronavirus Disease 2019 in China. The New England Journal of Medicine, 382(18), 1708–1720. doi:10.1056/NEJMoa2002032
Hamming, I., Timens, W., Bulthuis, M.L.C., Lely, A.T., Navis, G.J., & van Goor, H. (2004). Tissue Distribution of ACE2 Protein, the Functional Receptor for SARS Coronavirus. A First Step in Understanding SARS Pathogenesis. Journal of Pathology, 203(2), 631–637. doi:10.1002/path.1570
Higuchi, R., Fockler, C., Dollinger, G., & Watson, R. (1993). Kinetic PCR Analysis: Real-Time Monitoring of DNA Amplification Reactions. Biotechnology, 11(9), 1026-1030. doi:10.1038/nbt0993-1026
Hindson, J. (2020). COVID-19: faecal-oral transmission? Nature Reviews: Gastroenterology and Hepatology, 17(5), 259. doi:10.1038/s41575-020-0295-7
Jagodzinski, L.L., Manak, M.M., Hack, H.R., Liu, Y., & Peel, S.A. (2020). Performance evaluation of a laboratory developed PCR test for quantitation of HIV-2 viral RNA. PLoS One, 15(2), e0229424. doi:10.1371/journal.pone.0229424
Joshi, M., & Deshpande, J.D. (2011). Polymerase Chain Reaction: Methods, Principles and Application. International Journal of Biomedical Research, 2(1), 81-97. doi:10.7439/ijbr.v2i1.83
Kalifarhood, G., Aghaali, M., Saadati, H.M., Taherpour, N., Rahimi, S., Izadi, N., & Nazari, S.S.H. (2020). Epidemiological and Clinical Aspects of COVID-19; a Narrative Review. Archives of Academic Emergency Medicine, 8(1), e41.
van Kasteren, P.B., van der Veer, B., van den Brink, S., Wijsman, L., de Jonge, J., van den Brandt, A., Molenkamp, R., Reusken, C.B.E.M., & Meijer, A. (2020). Comparison of Seven Commercial RT-PCR Diagnostic Kits for COVID-19. Journal of Clinical Virology, 128, 104412. doi:10.1016/j.jcv.2020.104412
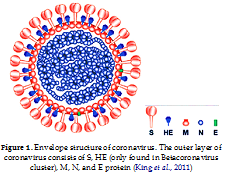
King, A.M.Q., Adams, M.J., Carstens, E.B., & Lefkowitz, E.J. (2011). Virus Taxonomy Ninth Report of the International Committee on Taxonomy of Viruses. Amsterdam, Netherlands: Elsevier B.V.
Kucirka, L.M., Lauer, S.A., Laeyendecker, O., Boon, D., & Lessler, J. (2020). Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction–Based SARS-CoV-2 Tests by Time Since Exposure. Annals of Internal Medicine, M20-1495. doi:10.7326/M20-1495
Lo, I.L., Lio, C.F., Cheong, H.H., Lei, C.I., Cheong, T.H., Zhong, X., Tian, Y., & Sin, N.N. (2020). Evaluation of SARS-CoV-2 RNA Shedding in Clinical Specimens and Clinical Characteristics of 10 Patients With COVID-19 in Macau. International Journal of Biological Sciences, 16(10), 1698–1707. doi:10.7150/ijbs.45357
Long, C., Xu, H., Shen, Q., Zhang, X., Fan, B., Wang, C., Zeng, B., Li, Z., Li, X., & Li, H. 2020. Diagnosis of the coronavirus disease (COVID-19): rRT-PCR or CT? European Journal of Radiology, 126, 108961. doi:10.1016/j.ejrad.2020.108961
Ministry of Health of the Republic of Indonesia. (2020a). Tak Ada Kasus nCoV Positif di Indonesia, Begini Alur Pemeriksaan Lab Balitbangkes. https://www.kemkes.go.id/article/view/20021200001/tak-ada-kasus-ncov-positif-di-indonesia-begini-alur-pemeriksaan-lab-balitbangkes.html
Ministry of Health of the Republic of Indonesia. (2020b). Pedoman Kesiapsiagaan Menghadapi Infeksi Novel Corona Virus (COVID-19) Revisi ke – 4. Jakarta, Indonesia: Directorate General of Disease Prevention and Control, Ministry of Health of the Republic of Indonesia.
Mullis, K.B. (1987). Process for amplifying nucleic acid sequences. United States Patent No. 4 683 202.
Pan, Y., Zhang, D., Yang, P., Poon, L.L.M., & Wang, Q. (2020). Viral Load of SARS-CoV-2 in Clinical Samples. The Lancet Infectious Diseases, 20(4), 411–412. doi:10.1016/S1473-3099(20)30113-4
Schaad, N.W., & Frederick, R.D. (2002). Real-time PCR and its application for rapid plant disease diagnostics. Canadian Journal of Plant Pathology, 24(3), 250–258. doi:10.1080/07060660209507006
Tahamtan, A. & Ardebili, A. (2020). Real-time RT-PCR in COVID-19 Detection: Issues Affecting the Results. Expert Review of Molecular Diagnostics, 20(5), 453–454. doi:10.1080/14737159.2020.1757437
Thai Public Broadcasting Service World. (2020). Machine error blamed for 40 false positive tests for COVID-19 in Yala. https://www.thaipbsworld.com/machine-error-blamed-for-40-false-positive-tests-for-covid-19-in-yala/
Wan, Y., Shang, J., Graham, R., Baric, R.S., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal of Virology, 94(7), 1–9. doi:10.1128/JVI.00127-20
Wang, W., Xu, Y., Gao, R., Lu, R., Han, K., Wu, G., & Tan, W. (2020). Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA, 323(18), 1843–1844. doi:10.1001/jama.2020.3786
Wölfel, R., Corman, V.M., Guggemos, W., Seilmaier, M., Zange, S., Müller, M.A., Niemeyer, D., Jones, T.C., Vollmar, P., Rothe, C., Hoelscher, M., Bleicker, T., Brünink, S., Schneider, J., Ehmann, R., Zwirglmaier, K., Drosten, C., & Wendtner, C. (2020). Virological Assessment of Hospitalized Patients With COVID-2019. Nature, 581(7809), 465-469. doi:10.1038/s41586-020-2196-x
World Health Organization. (2006). Collecting, Preserving and Shipping Specimens for The Diagnosis of Avian Influenza A(H5N1) virus infection. Guide for Field Operations. https://www.who.int/ihr/publications/Annex8.pdf?ua=1
World Health Organization. (2020a). Guidance for Laboratories Shipping Specimens to WHO Reference Laboratories That Provide Confirmatory Testing For COVID-19 Virus. https://apps.who.int/iris/bitstream/handle/10665/331639/WHO-2019-nCoV-laboratory_shipment-2020.3-eng.pdf
World Health Organization. (2020b). Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117
World Health Organization. (2020c). Interim Guidance: Clinical Management of Covid-19. https://www.who.int/publications-detail/clinical-management-of-covid-19
Worldometer. (2020). COVID-19 Coronavirus Pandemic. https://www.worldometers.info/coronavirus/
Zhang, L., Huang, Y., He, T., Cao, Y., & Ho, D. D. 1996. HIV-1 Subtype and Second-Receptor Use. Nature, 383(6603), 768. doi:10.1038/383768a0
Zou, L., Ruan, F., Huang, M., Liang, L., Huang, H., Hong, Z., Yu, J., Kang, M., Song, Y., Xia, J., Guo, Q., Song, T., He, J., Yen, H.L., Peiris, M., & Wu, J. (2020). SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. The New England Journal of Medicine, 382(12): 1175–1177. doi:10.1056/NEJMc2001737
Authors
Copyright (c) 2020 Maelita Ramdani Moeis, Anis Puji Rahayu, Nisa Ihsani, Wulan Pertiwi

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Authors continue to retain the copyright to the article if the article is published in the Borneo Journal of Pharmacy. They will also retain the publishing rights to the article without any restrictions.
Authors who publish in this journal agree to the following terms:
- Any article on the copyright is retained by the author(s).
- The author grants the journal the right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share work with an acknowledgment of the work authors and initial publications in this journal.
- Authors can enter into separate, additional contractual arrangements for the non-exclusive distribution of published articles (e.g., post-institutional repository) or publish them in a book, with acknowledgment of their initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their websites) prior to and during the submission process. This can lead to productive exchanges and earlier and greater citations of published work.
- The article and any associated published material are distributed under the Creative Commons Attribution-ShareAlike 4.0 International License.