Identification of Biological Risk Genes and Candidate Drugs for Psoriasis Vulgaris by Utilizing the Genomic Information
Abstract
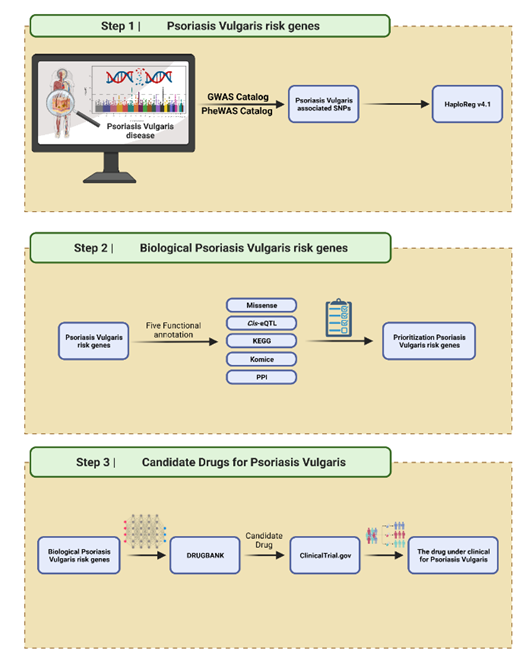
Psoriasis is an autoimmune disease that causes inflammation on the skin's surface, characterized by the appearance of pink plaques covered with white scales. Currently, the availability of psoriasis vulgaris therapy is still limited. Therefore, considering the discovery of new drug candidates by utilizing genetic variations, such as single nucleotide polymorphisms (SNP) through drug repurposing, is a profitable method. The SNP associated with psoriasis was obtained from Genome-Wide Association Studies (GWAS) and Phenom-Wide Association Studies (PheWAS) databases. We identified 245 SNPs associated with psoriasis vulgaris with criteria of r2 >0.8. To prioritize the candidate of a gene associated with psoriasis, we used five criteria of functional annotation (missense/nonsense, cis-eQTL, PPI, KEGG, and KO mice) where if there were more than two criteria of assessment, they were defined as the risk gene of psoriasis vulgaris. Fifty-two genes were identified as the risk gene of psoriasis vulgaris, then expanded using the STRING database to obtain more gene candidates of drug targets. The result is 104 genes candidates for drug targets, of which 24 overlapped with 96 drugs, according to DrugBank. Of the 96 drugs that have been approved for other indications, we found that five drugs (ustekinumab, tildrakizumab, risankizumab, guselkumab, and etanercept) are currently in clinical trials for the treatment of psoriasis that target two genes (IL23A and TNF). We argue that these two genes are the most promising targets based on their high target scores on functional annotations. This research explains the potential that utilizing genomic variation can contribute to drug discovery.
Full text article
References
2. Yang EJ, Beck KM, Sanchez IM, Koo J, Liao W. The impact of genital psoriasis on quality of life: a systematic review. Psoriasis. 2018;8:41-7. doi:10.2147/ptt.s169389
3. Yan BX, Chen XY, Ye LR, Chen JQ, Zheng M, Man XY. Cutaneous and Systemic Psoriasis: Classifications and Classification for the Distinction. Front Med. 2021;8:649408. doi:10.3389/fmed.2021.649408
4. Sarac G, Koca TT, Baglan T. A brief summary of clinical types of psoriasis. North Clin Istanb. 2016;3(1):79-82. doi:10.14744/nci.2016.16023
5. Singh RK, Lee KM, Ucmak D, Brodsky M, Atanelov Z, Farahnik B, et al. Erythrodermic psoriasis: pathophysiology and current treatment perspectives. Psoriasis. 2016;6:93-104. doi:10.2147/ptt.s101232
6. Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32:227-55. doi:10.1146/annurev-immunol-032713-120225
7. Burden AD, Javed S, Bailey M, Hodgins M, Conner M, Tillman D. Genetics of psoriasis: paternal inheritance and a locus on chromosome 6p. J Invest Dermatol. 1998;110(6):958–60. doi:10.1046/j.1523-1747.1998.00213.x
8. Sagoo GS, Tazi-Ahnini R, Barker JWN, Elder JT, Nair P, Samuelsson L, et al. Meta-analysis of genome-wide studies of psoriasis susceptibility reveals linkage to chromosomes 6p21 and 4q28-q31 in Caucasian and Chinese Hans population. J Invest Dermatol. 2004;122(6):1401–5. doi:10.1111/j.0022-202x.2004.22607.x
9. Trembath RC, Clough RL, Rosbotham, JL, Jones AB, Camp RD, Frodsham A, et al. Identification of a major susceptibility locus on chromosome 6p and evidence for further disease loci revealed by a two stage genome-wide search in psoriasis. Hum Mol Genet. 1997;6(5):813–20. doi:10.1093/hmg/6.5.813
10. Armstrong AW, Read C. Pathophysiology, Clinical Presentation, and Treatment of Psoriasis: A Review. JAMA. 2020;323(19):1945–60. doi:10.1001/jama.2020.4006
11. Kouris A, Platsidaki E, Kouskoukis C, Christodoulou C. Psychological parameters of psoriasis. Psychiatriki. 2017;28(1):54–9. doi:10.22365/jpsych.2017.281.54
12. Heath MS, Kolli SS, Dowling JR, Cline A, Feldman SR. Pharmacotherapeutic strategies for standard treatment-resistant psoriasis. Expert Opin Pharmacother. 2019;20(4):443–54. doi:10.1080/14656566.2018.1559819
13. Okwundu N, Cardwell LA, Cline AE, Richardson IM, Feldman SR. Adherence to topical treatment can improve treatment-resistant moderate psoriasis. Cutis. 2020;105(2):89-91;E2;E3.
14. Mohs RC, Greig NH. Drug discovery and development: Role of basic biological research. Alzheimers Dement. 2017;3(4):651-7. doi:10.1016/j.trci.2017.10.005
15. Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18(1):41–58. doi:10.1038/nrd.2018.168
16. Irham LM, Dania H, Maliza R, Faridah IN, Perwitasari DA. Pharmacogenomic : toward precision medicine. Yogyakarta: UAD Press; 2022.
17. Irham LM, Wong HSC, Chou WH, Adikusuma W, Mugiyanto E, Huang WC, et al. Integration of genetic variants and gene network for drug repurposing in colorectal cancer. Pharmacol Res. 2020;161:105203. doi:10.1016/j.phrs.2020.105203
18. Adikusuma W, Irham LM, Chou WH, Wong HSC, Mugiyanto E, Ting J, et al. Drug Repurposing for Atopic Dermatitis by Integration of Gene Networking and Genomic Information. Front Immunol. 2021;12:724277. doi:10.3389/fimmu.2021.724277
19. Bush WS, Moore JH. Chapter 11: Genome-wide association studies. PLoS Comput Biol. 2012;8(12):e1002822. doi:10.1371/journal.pcbi.1002822
20. Denny JC, Bastarache L, Roden DM. Phenome-Wide Association Studies as a Tool to Advance Precision Medicine. Annu Rev Genomics Hum Genet. 2016;17:353-73. doi:10.1146/annurev-genom-090314-024956
21. Lesmana MHS, Le NQK, Chiu WC, Chung KH, Wang CY, Irham LM, et al. Genomic-Analysis-Oriented Drug Repurposing in the Search for Novel Antidepressants. Biomedicines. 2022;10(8):1947. doi:10.3390/biomedicines10081947
22. Adikusuma W, Chou WH, Lin MR, Ting J, Irham LM, Perwitasari DA, et al. Identification of Druggable Genes for Asthma by Integrated Genomic Network Analysis. Biomedicines. 2022;10(1):113. doi:10.3390/biomedicines10010113
23. Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901-6. doi:10.1093/nar/gkm958
24. Guharoy V. Clinicaltrials.gov: Is the Glass Half Full? Hosp Pharm. 2014;49(10):893-5. doi:10.1310/hpj4910-893
25. Aggeletopoulou I, Assimakopoulos SF, Konstantakis C, Triantos C. Interleukin 12/interleukin 23 pathway: Biological basis and therapeutic effect in patients with Crohn's disease. World J Gastroenterol. 2018;24(36):4093-103. doi:10.3748/wjg.v24.i36.4093
26. Langrish CL, McKenzie BS, Wilson NJ, Malefyt RdW, Kastelein RA, Cua DJ. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol Rev. 2004;202:96-105. doi:10.1111/j.0105-2896.2004.00214.x
27. Tang C, Chen S, Qian H, Huang W. Interleukin-23: as a drug target for autoimmune inflammatory diseases. Immunology. 2012;135(2):112-24. doi:10.1111/j.1365-2567.2011.03522.x
28. Sinclair R, Palanivelu VT. Tildrakizumab for the treatment of psoriasis. Expert Rev Clin Immunol. 2019;15(1):5–12. doi:10.1080/1744666x.2019.1544493
29. Deodhar A, Helliwell PS, Boehncke WH, Kollmeier AP, Hsia EC, Subramanian RA, et al. Guselkumab in patients with active psoriatic arthritis who were biologic-naive or had previously received TNFα inhibitor treatment (DISCOVER-1): a double-blind, randomised, placebo-controlled phase 3 trial. Lancet. 2020;395(10230):1115–25. doi:10.1016/s0140-6736(20)30265-8
30. Wang X, Lin Y. Tumor Necrosis Factor and cancer, buddies or foes? Acta Pharmacol Sin. 2008;29(11):1275–88. doi:10.1111/j.1745-7254.2008.00889.x
31. Campa M, Ryan C, Menter A. An overview of developing TNF-α targeted therapy for the treatment of psoriasis. Expert Opin Investig Drugs. 2015;24(10):1343–54. doi:10.1517/13543784.2015.1076793
32. Li SJ, Perez-Chada LM, Merola JF. TNF Inhibitor-Induced Psoriasis: Proposed Algorithm for Treatment and Management. J Psoriasis Psoriatic Arthritis. 2019;4(2):70–80. doi:10.1177/2475530318810851
33. Jang DI, Lee AH, Shin HY, Song HR, Park JH, Kang TB, et al. The Role of Tumor Necrosis Factor Alpha (TNF-α) in Autoimmune Disease and Current TNF-α Inhibitors in Therapeutics. Int J Mol Sci. 2021;22(5):2719. doi:10.3390/ijms22052719
34. Steeland S, Libert C, Vandenbroucke RE. A New Venue of TNF Targeting. Int J Mol Sci. 2018;19(5):1442. doi:10.3390/ijms19051442
35. Zhou H. Clinical pharmacokinetics of etanercept: A fully humanized soluble recombinant Tumor Necrosis Factor receptor fusion protein. J Clin Pharmacol. 2005;45(5):490–7. doi:10.1177/0091270004273321
36. Liang S, Dai J, Hou S, Su L, Zhang D, Guo H, et al. Structural basis for treating tumor necrosis factor α (TNFα)-associated diseases with the therapeutic antibody infliximab. J Biol Chem. 2013;288(19):13799-807. doi:10.1074/jbc.m112.433961
37. Subedi S, Gong Y, Chen Y, Shi Y. Infliximab and biosimilar infliximab in psoriasis: efficacy, loss of efficacy, and adverse events. Drug Des Devel Ther. 2019;13:2491-502. doi:10.2147/dddt.s200147
38. Nast A, Sporbeck B, Rosumeck S, Pathirana D, Jacobs A, Werner RN, et al. Which antipsoriatic drug has the fastest onset of action?-systematic review on the rapidity of the onset of action. J Invest Dermatol. 2013;133(8):1963–70. doi:10.1038/jid.2013.78
39. de Vries ACQ, Thio HB, de Kort WJA, Opmeer BC, van der Stok HM, de Jong EMGJ, et al. A prospective randomized controlled trial comparing infliximab and etanercept in patients with moderate-to-severe chronic plaque-type psoriasis: the Psoriasis Infliximab vs. Etanercept Comparison Evaluation (PIECE) study. Br J Dermatol. 2017;176(3):624–33. doi:10.1111/bjd.14867
Authors
Copyright (c) 2023 Lisza Niarisessa, Anisa Nova Puspitaningrum, Arief Rahman Afief, Dyah Aryani Perwitasari, Wirawan Adikusuma, Rocky Cheung, Abdi Wira Septama, Lalu Muhammad Irham

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Authors continue to retain the copyright to the article if the article is published in the Borneo Journal of Pharmacy. They will also retain the publishing rights to the article without any restrictions.
Authors who publish in this journal agree to the following terms:
- Any article on the copyright is retained by the author(s).
- The author grants the journal the right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share work with an acknowledgment of the work authors and initial publications in this journal.
- Authors can enter into separate, additional contractual arrangements for the non-exclusive distribution of published articles (e.g., post-institutional repository) or publish them in a book, with acknowledgment of their initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their websites) prior to and during the submission process. This can lead to productive exchanges and earlier and greater citations of published work.
- The article and any associated published material are distributed under the Creative Commons Attribution-ShareAlike 4.0 International License.