Molecular Docking and Dynamics of Xylocarpus granatum as A Potential Parkinson’s Drug Targeting Multiple Enzymes
Abstract
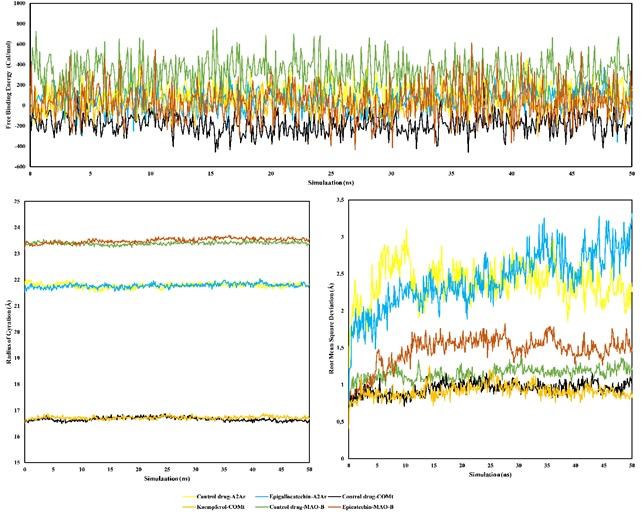
Parkinson's disease is a global health challenge affecting over 10 million individuals worldwide, leading to increased disability-adjusted life years (DALYs) and a rise in mortality rates. This study explores the potential anti-Parkinson's properties of Xylocarpus granatum, focusing on its interaction with key enzymes associated with the disease: catechol-O-methyltransferase (COMT), adenosine A2A receptor (A2AR), and monoamine oxidase-B (MAO-B). Using molecular docking and molecular dynamics approaches with YASARA Structure, the ethanol extract of X. granatum was investigated for its mechanism of action. Among 30 compounds, five demonstrated promising binding affinities. Structural flexibility analysis revealed minimal fluctuations in active-site residues, highlighting the stability of key complexes involving kaempferol, epicatechin, epigallocatechin, and native ligands. Molecular Mechanics Poisson–Boltzmann Surface Area (MM-PBSA) simulations provided insights into the binding energy of these complexes. Notably, kaempferol exhibited higher binding energy than the natural ligand, suggesting superior binding affinity. Analysis of the average radius of gyration (Rg) showcased control drug-MAO-B exhibited higher Rg values, indicating a more flexible protein conformation. Confirming mode stability with root mean square deviation (RMSD) analysis shows overall stability, except in the A2AR-bound complex. The study's collective findings underscore the structural stabilization of ligand-protein complexes, contributing valuable insights into the potential anti-Parkinson's properties of X. granatum. These discoveries hold promise for developing more effective therapies for Parkinson's disease and significantly contribute to the neurology field.
Full text article
References
2. Kouli A, Torsney KM, Kuan WL. Parkinson’s Disease: Etiology, Neuropathology, and Pathogenesis. In: Stoker TB, Greenland JC, editors. Parkinson’s Disease: Pathogenesis and Clinical Aspects. Brisbane (AU): Codon Publications; 2018. DOI: 10.15586/codonpublications.parkinsonsdisease.2018.ch1
3. DeMaagd G, Philip A. Parkinson's Disease and Its Management: Part 1: Disease Entity, Risk Factors, Pathophysiology, Clinical Presentation, and Diagnosis. P T. 2015;40(8):504-32. PMCID: PMC4517533; PMID: 26236139
4. Magrinelli F, Picelli A, Tocco P, Federico A, Roncari L, Smania N, et al. Pathophysiology of Motor Dysfunction in Parkinson's Disease as the Rationale for Drug Treatment and Rehabilitation. Parkinsons Dis. 2016;2016:9832839. DOI: 10.1155/2016/9832839; PMCID: PMC4913065; PMID: 27366343
5. Jankovic J, Tan EK. Parkinson’s disease: etiopathogenesis and treatment. J Neurol Neurosurg Psychiatry. 2020;91(8):795-808. DOI: 10.1136/jnnp-2019-322338; PMID: 32576618
6. Regensburger M, Ip CW, Kohl Z, Schrader C, Urban PP, Kassubek J, et al. Clinical benefit of MAO-B and COMT inhibition in Parkinson’s disease: practical considerations. J Neural Transm. 2023;130(6):847-61. DOI: 10.1007/s00702-023-02623-8; PMCID: PMC10199833; PMID: 36964457
7. Boulaamane Y, Ibrahim MAA, Britel MR, Maurady A. In silico studies of natural product-like caffeine derivatives as potential MAO-B inhibitors/AA 2A R antagonists for the treatment of Parkinson’s disease. J Integr Bioinform. 2022;19(4):20210027. DOI: 10.1515/jib-2021-0027; PMCID: PMC9800045; PMID: 36112816
8. Dey D, Quispe C, Hossain R, Jain D, Ahmed Khan R, Janmeda P, et al. Ethnomedicinal Use, Phytochemistry, and Pharmacology of Xylocarpus granatum J. Koenig. Emran T Bin, editor. Evid Based Complement Altern Med. 2021;2021:8922196. DOI: 10.1155/2021/8922196; PMCID: PMC8423563; PMID: 34504541
9. Yin X, Li X, Hao Y, Zhao Y, Zhou J, Shi H. Xylocarpin H, a Limonoid of Xylocarpus granatum, Produces Antidepressant-Like Activities in Mice. J Behav Brain Sci. 2015;5(11):524-32. DOI: 10.4236/jbbs.2015.511050
10. Agu PC, Afiukwa CA, Orji OU, Ezeh EM, Ofoke IH, Ogbu CO, et al. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci Rep. 2023;13(1):13398. DOI: 10.1038/s41598-023-40160-2; PMCID: PMC10435576; PMID: 37592012
11. Hollingsworth SA, Dror RO. Molecular Dynamics Simulation for All. Neuron. 2018;99(6):1129-43. DOI: 10.1016/j.neuron.2018.08.011; PMCID: PMC6209097; PMID: 30236283
12. Heryanto R, Putra CA, Khalil M, Rafi M, Putri SP, Karomah AH, et al. Antioxidant Activity and Metabolite Profiling of Xylocarpus granatum Extracts Using Gas Chromatography-Mass Spectrometry. Metabolites. 2023;13(2):156. DOI: 10.3390/metabo13020156; PMCID: PMC9958973; PMID: 36837775
13. Irsal RAP, Seno DSH, Safithri M, Kurniasih R. Penapisan Virtual Senyawa Aktif Sirih Merah (Piper Crocatum) sebagai Inhibitor Angiotensin Converting Enzyme. J Farmamedika Pharmamedika J. 2022;7(2):104-13. DOI: 10.47219/ath.v7i2.157
14. Durai P, Shin H, Achek A, Kwon H, Govindaraj RG, Panneerselvam S, et al. Toll‐like receptor 2 antagonists identified through virtual screening and experimental validation. FEBS J. 2017;284(14):2264-83. DOI: 10.1111/febs.14124; PMID: 28570013
15. Gholam GM, Darmawan NI, Siregar JE, Artika IM. Selected Polyphenols from Date (Phoenix dactylifera) as Anti-Virulence of Candida albicans Through Multiple Enzyme Targets. Biointerface Res Appl Chem. 2022;13(4):386. DOI: 10.33263/BRIAC134.386
16. Talmaciu MM, Bodoki E, Oprean R. Global chemical reactivity parameters for several chiral beta-blockers from the Density Functional Theory viewpoint. Clujul Med. 2016;89(4):513-8. DOI: 10.15386/cjmed-610; PMCID: PMC5111492; PMID: 27857521
17. Land H, Humble MS. YASARA: A Tool to Obtain Structural Guidance in Biocatalytic Investigations. In: Bornscheuer UT, Höhne M, editors. Protein Engineering. Methods in Molecular Biology. New York (US): Humana Press: 2018. p. 43–67. DOI: 10.1007/978-1-4939-7366-8_4
18. Chairunisa F, Safithri M, Andrianto D, Kurniasih R, Irsal RAP. Molecular Docking of Red Betel Leaf Bioactive Compounds (Piper crocatum) as Lipoxygenase Inhibitor. Indones J Pharm Sci Technol. 2023;10(2):90-103. DOI: 10.24198/ijpst.v10i2.38934
19. Odhar HA, Hashim AF, Humad SS. Molecular docking analysis and dynamics simulation of salbutamol with the monoamine oxidase B (MAO-B) enzyme. Bioinformation. 2022;18(3):304-9. DOI: 10.6026/97320630018304; PMCID: PMC9722423; PMID: 36518132
20. Ferenczy GG, Kellermayer M. Contribution of hydrophobic interactions to protein mechanical stability. Comput Struct Biotechnol J. 2022;20:1946-56. DOI: 10.1016/j.csbj.2022.04.025; PMCID: PMC9062142; PMID: 35521554
21. Dhorajiwala TM, Halder ST, Samant L. Computer-Aided Docking Studies of Phytochemicals from Plants Salix Subserrata and Onion as Inhibitors of Glycoprotein G of Rabies Virus. Biomed Biotechnol Res J. 2019;3(4):269-76. DOI: 10.4103/bbrj.bbrj_124_19
22. Roohi H, Mohtamadifar N. The role of the donor group and electron-accepting substitutions inserted in π-linkers in tuning the optoelectronic properties of D-π-A dye-sensitized solar cells: a DFT/TDDFT study. RSC Adv. 2022;12(18):11557-73. DOI: 10.1039/d2ra00906d; PMCID: PMC9006569; PMID: 35425060
23. Chattaraj PK, Duley S. Electron Affinity, Electronegativity, and Electrophilicity of Atoms and Ions†. J Chem Eng Data. 2010;55(5):1882-6. DOI: 10.1021/je900892p
24. Zhan CG, Nichols JA, Dixon DA. Ionization Potential, Electron Affinity, Electronegativity, Hardness, and Electron Excitation Energy: Molecular Properties from Density Functional Theory Orbital Energies. J Phys Chem A. 2003;107(20):4184-95. DOI: 10.1021/jp0225774
25. Dash R, Ali MC, Dash N, Azad MAK, Hosen SMZ, Hannan MA, et al. Structural and Dynamic Characterizations Highlight the Deleterious Role of SULT1A1 R213H Polymorphism in Substrate Binding. Int J Mol Sci. 2019;20(24):6256. DOI: 10.3390/ijms20246256; PMCID: PMC6969939; PMID: 31835852
26. Biswas S, Mahmud S, Mita MA, Afrose S, Hasan MR, Shimu MSS, et al. Molecular Docking and Dynamics Studies to Explore Effective Inhibitory Peptides Against the Spike Receptor Binding Domain of SARS-CoV-2. Front Mol Biosci. 2022;8(10):1305-15. DOI: 10.3389/fmolb.2021.791642; PMCID: PMC8851422; PMID: 35187069
27. Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10(5):449-61. DOI: 10.1517/17460441.2015.1032936; PMCID: PMC4487606; PMID: 25835573
28. Justino GC, Nascimento CP, Justino MC. Molecular dynamics simulations and analysis for bioinformatics undergraduate students. Biochem Mol Biol Educ. 2021;49(4):570-82. DOI: 10.1002/bmb.21512; PMID: 33844418
29. Ng HW, Laughton CA, Doughty SW. Molecular dynamics simulations of the adenosine A2a receptor: structural stability, sampling, and convergence. J Chem Inf Model. 2013;53(5):1168-78. DOI: 10.1021/ci300610w; PMID: 23514445
Authors
Copyright (c) 2024 Riyan Alifbi Putera Irsal, Gusnia Meilin Gholam, Dzikri Anfasa Firdaus, Novian Liwanda, Fernanda Chairunisa

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Authors continue to retain the copyright to the article if the article is published in the Borneo Journal of Pharmacy. They will also retain the publishing rights to the article without any restrictions.
Authors who publish in this journal agree to the following terms:
- Any article on the copyright is retained by the author(s).
- The author grants the journal the right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share work with an acknowledgment of the work authors and initial publications in this journal.
- Authors can enter into separate, additional contractual arrangements for the non-exclusive distribution of published articles (e.g., post-institutional repository) or publish them in a book, with acknowledgment of their initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their websites) prior to and during the submission process. This can lead to productive exchanges and earlier and greater citations of published work.
- The article and any associated published material are distributed under the Creative Commons Attribution-ShareAlike 4.0 International License.