Detection of Salmonella typhimurium ATCC 14028 in Powder Prepared Traditional Medicines Using Real-Time PCR
Abstract
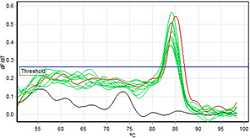
The detection of Salmonella typhimurium ATCC 14028 using real-time PCR on powdered traditional medicinal products was carried out in the microbiology and molecular biology testing laboratory of the Food and Drug Administration in Gorontalo. This research aims to provide a reference for alternative testing methods in testing the products of traditional powder preparations on the market. The sample consisted of 10 traditional powder preparations spiked with positive control of S. typhimurium ATCC 14028 phase 2. The method used in the study was real-time PCR analysis using the SYBR® Green method, while DNA isolation using the direct PCR method. Data analysis was performed by analyzing the sample's melting temperature (Tm) curve and comparing it with positive control. The results showed that S. typhimurium ATCC 14028 was detected in samples at an average Tm value of 84.18°C, with ranges of 84.0-84.5°C. For positive control, the Tm value was at 85.2°C, while for the negative control, the Tm value was not detected. Based on these data, it can be concluded that S. typhimurium ATCC 14028 in traditional medicine products powder preparations can be detected using real-time PCR.
Full text article
References
2. Sidoretno WM, Rz IO. Edukasi Bahaya Bahan Kimia Obat Yang Terdapat Didalam Obat Tradisional. Jurnal Pengabdian Masyarakat Multidisiplin. 2018;1(2):36-42. doi:10.36341/jpm.v1i2.453
3. Abba D, Inabo HI, Yakubu SE, Olonitola OS. Contamination of herbal medicinal products marketed in Kaduna metropolis with selected pathogenic bacteria. Afr J Tradit Complement Altern Med. 2008;6(1):70-7. doi:10.4314/ajtcam.v6i1.57076
4. Kumadoh D, Ofori-Kwakye K. Dosage Forms of Herbal Medicinal Products and Their Stability Considerations-an Overview. J Crit Rev. 2017;4(4):1-8. doi:10.22159/jcr.2017v4i4.16077
5. Franco-Duarte R, Černáková L, Kadam S, Kaushik KS, Salehi B, Bevilacqua A, et al. Advances in Chemical and Biological Methods to Identify Microorganisms-From Past to Present. Microoganisms. 2019;7(5):130. doi:10.3390/microorganisms7050130
6. Law JWF, Ab Mutalib NS, Chan KG, Lee LH. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol. 2015;5:770. doi:10.3389/fmicb.2014.00770
7. Maurer FP, Christner M, Hentschke M, Rohde H. Advances in Rapid Identification and Susceptibility Testing of Bacteria in the Clinical Microbiology Laboratory: Implications for Patient Care and Antimicrobial Stewardship Programs. Infect Dis Rep. 2017;9(1):6839. doi:10.4081/idr.2017.6839
8. Priyanka B, Patil RK, Dwarakanath S. A review on detection methods used for foodborne pathogens. Indian J Med Res. 2016;144(3):327-38. doi:10.4103/0971-5916.198677
9. Bintsis T. Foodborne pathogens. AIMS Microbiol. 2017;3(3):529-63. doi:10.3934/microbiol.2017.3.529
10. Jajere SM. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet World. 2019;12(4):504-21. doi:10.14202/vetworld.2019.504-521
11. Kasturi KN, Drgon T. Real-Time PCR Method for Detection of Salmonella spp. in Environmental Samples. Appl Environ Microbiol. 2017;83(14):e00644-17. doi:10.1128/AEM.00644-17
12. Ekor M. The growing use of herbal medicines: issues relating to adverse reactions and challenges in monitoring safety. Front Pharmacol. 2014;4:177. doi:10.3389/fphar.2013.00177
13. Sophian A, Purwaningsih R, Igirisa EPJ, Amirullah MA, Lukita BL, Fitri RA. Short Communication: Detection of Salmonella typhimurium ATCC 14028 and Listeria monocytogenes ATCC 7644 in processed meat products using Real-Time PCR Multiplex Method. Asian J Nat Prod Biochem. 2021;19(1):17-20. doi:10.13057/biofar/f190103
14. Sophian A, Purwaningsih R, Lukita BL, Ningsih EC. Detection of Salmonella typhimurium ATCC 14028 in supplement health product liquid preparation using Real-Time PCR (qPCR). Asian J Nat Prod Biochem. 2020;18(2):61-65. doi:10.13057/biofar/f180202
15. Kang DH, Fung DY. Application of thin agar layer method for recovery of injured Salmonella typhimurium. Int J Food Microbiol. 2000;54(1-2):127-32. doi:10.1016/s0168-1605(99)00174-9
16. Lagier JC, Edouard S, Pagnier I, Mediannikov O, Drancourt M, Raoult D. Current and Past Strategies for Bacterial Culture in Clinical Microbiology. Clin Microbiol Rev. 2015;28(1):208-36. doi:10.1128/CMR.00110-14
17. Bonnet M, Lagier JC, Raoult D, Khelaifia S. Bacterial culture through selective and non-selective conditions: the evolution of culture media in clinical microbiology. New Microbes New Infect. 2020;34:100622. doi:10.1016/j.nmni.2019.100622
18. Park SH, Ryu S, Kang DH. Development of an Improved Selective and Differential Medium for Isolation of Salmonella spp. J Clin Microbiol. 2012;50(10):3222-6. doi:10.1128/JCM.01228-12
19. El Shamy HA, Bakr WM, Gomaa NF, Barheem OH. Evaluation of two enrichment broths, three plating media and ELISA technique for the isolation of salmonella from dairy products. J Egypt Public Health Assoc. 2008;83(1-2):133-45.
20. Gupta N. DNA Extraction and Polymerase Chain Reaction. J Cytol. 2019;36(2):116-7. doi:10.4103/JOC.JOC_110_18
21. Shinozuka H, Forster JW. Use of the melting curve assay as a means for high-throughput quantification of Illumina sequencing libraries. PeerJ. 2016;4:e2281. doi:10.7717/peerj.2281
22. Zhang H, Gaňová M, Yan ZQ, Chang H, Neužil P. PCR Multiplexing Based on a Single Fluorescent Channel Using Dynamic Melting Curve Analysis. ACS Omega. 2020;5(46):30267-73. doi:10.1021/acsomega.0c04766
23. Ali N, Rampazzo RdCP, Costa ADT, Krieger MA. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. Biomed Res Int. 2017;2017:9306564. doi:10.1155/2017/9306564
24. Kralik P, Ricchi M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front Microbiol. 2017;8:108. doi:10.3389/fmicb.2017.00108
25. Gašparič MB, Cankar K, Žel J, Gruden K. Comparison of different real-time PCR chemistries and their suitability for detection and quantification of genetically modified organisms. BMC Biotechnol. 2008;8:26. doi:10.1186/1472-6750-8-26
Authors
Copyright (c) 2021 Alfi Sophian, Ratna Purwaningsih, Muindar Muindar, Eka Putri Juniarti Igirisa, Muhammad Luthfi Amirullah

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Authors continue to retain the copyright to the article if the article is published in the Borneo Journal of Pharmacy. They will also retain the publishing rights to the article without any restrictions.
Authors who publish in this journal agree to the following terms:
- Any article on the copyright is retained by the author(s).
- The author grants the journal the right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share work with an acknowledgment of the work authors and initial publications in this journal.
- Authors can enter into separate, additional contractual arrangements for the non-exclusive distribution of published articles (e.g., post-institutional repository) or publish them in a book, with acknowledgment of their initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their websites) prior to and during the submission process. This can lead to productive exchanges and earlier and greater citations of published work.
- The article and any associated published material are distributed under the Creative Commons Attribution-ShareAlike 4.0 International License.