Use of Direct PCR Technique Without DNA Extraction in Confirmation Test for Salmonella typhimurium Bacteria on Meatball Samples
Abstract
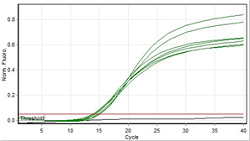
The use of direct PCR technique without DNA extraction in the confirmation test for Salmonella typhimurium ATCC 14028 bacteria on meatball samples was carried out in the Food and Drug molecular biology testing laboratory Administration in Gorontalo. The basis of this research is to have an impact on economic value in carrying out the confirmation test for S. typhimurium ATCC 14028, where testing is carried out conventionally, namely DNA extraction, which requires a large amount of money. Hence, it is necessary to innovate to modify the testing phase so that it is more effective and efficient. The purpose of this study was to see whether the direct PCR technique without DNA extraction can be done for the confirmation test of S. typhimurium ATCC 14028 on meatball samples. This study's sample consisted of 20 types of meatball samples spiked with S. typhimurium ATCC 14028 cultures. The method used in this study was qPCR analysis using the SYBR Green method. Data analysis was carried out based on 2 main criteria: (1) Ct analysis and (2) Tm analysis. Real-time PCR analysis results obtained Ct values in the range 14.14 - 15.20 with an average of 14.82 and Tm values 85.20 - 86.30 with an average of 85.79. Based on these data, it can be concluded that using direct PCR can be used for testing confirmation of S. typhimurium ATCC 14028 on meatball samples.
Full text article
References
2. Kotsiri Z, Vantarakis A, Rizzotto F, Kavanaugh D, Ramarao N, Vidic J. Sensitive Detection of E. coli in Artificial Seawater by Aptamer-Coated Magnetic Beads and Direct PCR. Appl Sci. 2019;9(24):5392. doi:10.3390/app9245392
3. Aymerich T, Martín B, Garriga M, Hugas M. Microbial quality and direct PCR identification of lactic acid bacteria and nonpathogenic Staphylococci from artisanal low-acid sausages. Appl Environ Microbiol. 2003;69(8):4583-94. doi:10.1128/aem.69.8.4583-4594.2003
4. Morecchiato F, Coppi M, Baccani I, Maggini N, Ciccone N, Antonelli A, et al. Evaluation of extraction-free RT-PCR methods for faster and cheaper detection of SARS-CoV-2 using two commercial systems. Int J Infect Dis. 2021;112:264-8. doi:10.1016/j.ijid.2021.09.046
5. Xiao L, Zhang Z, Sun X, Pan Y, Zhao Y. Development of a quantitative real-time PCR assay for viable Salmonella spp. without enrichment. Food Control. 2015;57:185-9. doi:10.1016/j.foodcont.2015.03.050
6. Miller ND, Davidson PM, D’Souza DH. Real-time reverse-transcriptase PCR for Salmonella Typhimurium detection from lettuce and tomatoes. LWT - Food Sci Technol. 2011;44(4):1088-97. doi:10.1016/j.lwt.2010.08.003
7. D'Urso OF, Poltronieri P, Marsigliante S, Storelli C, Hernández M, Rodríguez-Lázaro D. A filtration-based real-time PCR method for the quantitative detection of viable Salmonella enterica and Listeria monocytogenes in food samples. Food Microbiol. 2009;26(3):311-6. doi:10.1016/j.fm.2008.12.006
8. Elizaquível P, Aznar R. A multiplex RTi-PCR reaction for simultaneous detection of Escherichia coli O157:H7, Salmonella spp. and Staphylococcus aureus on fresh, minimally processed vegetables. Food Microbiol. 2008;25(5):703-13. doi:10.1016/j.fm.2008.03.002
9. Andrews JR, Ryan ET. Diagnostics for invasive Salmonella infections: current challenges and future directions. Vaccine. 2015;33(Suppl 3):C8-C15. doi:10.1016/j.vaccine.2015.02.030
10. Ranjbar R, Mortazavi SM, Tavana AM, Sarshar M, Najafi A, Zanjani RS. Simultaneous Molecular Detection of Salmonella enterica Serovars Typhi, Enteritidis, Infantis, and Typhimurium. Iran J Public Health. 2017;46(1):103-11.
11. Yang S, Rothman RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. 2004;4(6):337-48. doi:10.1016/s1473-3099(04)01044-8
12. Holt JG, Krieg NR, Sneath PHA, Stanley JT, William ST. Bergey’s Manual of Determinative Bacteriology. 9th edition. Baltimore (MD): Lippincott Williams & Wilkins; 2000.
13. Ramachandran G. Gram-positive and gram-negative bacterial toxins in sepsis. Virulence. 2014;5(1):213-8. doi:10.4161/viru.27024
14. Bell RL, Jarvis KG, Ottesen AR, McFarland MA, Brown EW. Recent and emerging innovations in Salmonella detection: a food and environmental perspective. Microb Biotechnol. 2016;9(3):279-92. doi:10.1111/1751-7915.12359
15. Adzitey F, Huda N, Ali GRR. Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. 3 Biotech. 2013;3(2):97-107. doi:10.1007/s13205-012-0074-4
16. Postollec F, Falentin H, Pavan S, Combrisson J, Sohier D. Recent advances in quantitative PCR (qPCR) applications in food microbiology. Food Microbiol. 2011;28(5):848-61. doi:10.1016/j.fm.2011.02.008
17. Beutin L, Jahn S, Fach P. Evaluation of the 'GeneDisc' real-time PCR system for detection of enterohaemorrhagic Escherichia coli (EHEC) O26, O103, O111, O145 and O157 strains according to their virulence markers and their O- and H-antigen-associated genes. J Appl Microbiol. 2009;106(4):1122-32. doi:10.1111/j.1365-2672.2008.04076.x
18. Botteldoorn N, Heyndrickx M, Rijpens N, Herman L. Detection and characterization of verotoxigenic Escherichia coli by a VTEC/EHEC multiplex PCR in porcine faeces and pig carcass swabs. Res Microbiol. 2003;154(2):97-104. doi:10.1016/s0923-2508(03)00028-7
19. Fukushima H, Katsube K, Tsunomori Y, Kishi R, Atsuta J, Akiba Y. Comprehensive and rapid real-time PCR analysis of 21 foodborne outbreaks. Int J Microbiol. 2009;2009:917623. doi:10.1155/2009/917623
20. Kim HJ, Cho JC. Simple and rapid detection of Listeria monocytogenes in fruit juice by real-time PCR without enrichment culture. Food Control. 2010;21(10):1419-23. doi:10.1016/j.foodcont.2010.04.006
21. Singh J, Batish VK, Grover S. Simultaneous detection of Listeria monocytogenes and Salmonella spp. in dairy products using real time PCR-melt curve analysis. J Food Sci technol. 2012;49(2):234-9. doi:10.1007/s13197-011-0278-3
22. Rose BE, Hill WE, Umholtz R, Ransom GM, James WO. Testing for Salmonella in raw meat and poultry products collected at federally inspected establishments in the United States, 1998 through 2000. J Food Prot. 2002;65(6):937-47. doi:10.4315/0362-028x-65.6.937
23. Zain R, Hidanah S, Damayanti R, Warsito SH. Detection of Salmonella sp. on bulk meatballs and packaged meatballs at Sepanjang Market, Sidoarjo. J Appl Vet Sci Technol. 2021;2(2):31-6. doi:10.20473/javest.V2.I2.2021.31-36
24. Sophian A, Muindar. Pengujian Salmonella typhimurium ATCC 14028 pada produk sosis, nugget, bakso, otak-otak, tempura dan cilok menggunakan kit rapid test. J Exp Clin Pharm. 2021;1(1):48-55. doi:10.52365/jecp.v1i1.198
25. Sophian A, Purwaningsih R, Igirisa EPJ, Amirullah ML, Lukita BL, Fitri RA. Short Communication: Detection of Salmonella typhimurium ATCC 14028 and Listeria monocytogenes ATCC 7644 in processed meat products using Real-Time PCR Multiplex Method. Asian J Nat Prod Biochem. 2021;19(1):17-20. doi:10.13057/biofar/f190103
26. Sophian A, Purwaningsih R, Muindar, Igirisa EPJ, Amirullah ML. Detection of Salmonella typhimurium ATCC 14028 in Powder Prepared Traditional Medicines Using Real-Time PCR. Borneo J Pharm. 2021;4(3):178-83. doi:10.33084/bjop.v4i3.1838
27. Sophian A, Purwaningsih R, Lukita BL, Ningsih EC. Detection of Salmonella typhimurium ATCC 14028 in supplement health product liquid preparation using Real-Time PCR (qPCR). Asian J Nat Prod Biochem. 2020;18(2):65-6. doi:10.13057/biofar/f180202
28. Sapiun Z, Kamba V, Damiti SA, Sophian A, Abinawanto, Muindar, et al. Optimization of McFarland Turbidity Standards Value in Determining Template DNA as Reference in Salmonella typhimurium ATCC 14028 Test Using Real-Time PCR (qPCR). PalArch's J Archaeol Egypt/ Egyptol. 2020;17(6):10916-21.
29. Andrews WH, Wang H, Jacobson A, Ge B, Zhang G, Hammack T. Bacteriological Analytical Manual. BAM Chapter 5: Salmonella. Silver Spring (MD): The United States Food and Drug Administration; 2007 [updated 2021 Oct; cited 2021 Nov]. Available from: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-5-salmonella
30. Baylis CL, MacPhee S, Betts RP. Comparison of two commercial preparations of buffered peptone water for the recovery and growth of Salmonella bacteria from foods. J Appl Microbiol. 2000;89(3):501-10. doi:10.1046/j.1365-2672.2000.01145.x
31. Gupta N. DNA Extraction and Polymerase Chain Reaction. J Cytol. 2019;36(2):116-7. doi:10.4103/JOC.JOC_110_18
32. Nugraha R, Nurilmala M, Nurjanah, Pratama P. Detection of Salmonella sp. in fisheries product using real-time PCR. IOP Conf Ser Earth Environ Sci. 2020;404:012012. doi:10.1088/1755-1315/404/1/012012
33. de Oliveira ACdS, Rosa MC, Bochardt JL, Menegon YA, Fernandes MMA, Cardoso GVF, et al. Validating the Efficiency of a Simplex PCR and Quantitative SYBR Green qPCR for the Identification of Salmonella spp. DNA. J Food Microbiol Saf Hyg. 2018;3:1. doi:10.4172/2476-2059.1000130
34. Dorak M. Real-time PCR. London (UK): Taylor & Francis; 2006. doi:10.4324/9780203967317
35. Karlen Y, McNair A, Perseguers S, Mazza C, Mermod N. Statistical significance of quantitative PCR. BMC Bioinformatics. 2007;8:131. doi:10.1186/1471-2105-8-131
36. Abtahi H, Sadeghi MR, Shabani M, Edalatkhah H, Hadavi R, Akhondi MM, et al. Causes of bimodal melting curve:Asymmetric guaninecytosine (GC) distribution causing two peaks in melting curve and affecting their shapes. Afr J Biotechnol. 2011;10(50:10196-203. doi:10.5897/AJB11.1782
37. Carter I, Halliday C, Sloots TP, Pryce TM, Kay ID, Harnett GB, et al. PCR Methodology. In: Schuller M, Sloots T, James G, Halliday C, Carter I, editors. PCR for Clinical Microbiology. Dordrecht: Springer; 2010. p. 11-47. doi:10.1007/978-90-481-9039-3_2
38. Lorenz TC. Polymerase Chain Reaction: Basic Protocol Plus Troubleshooting and Optimization Strategies. J Vis Exp. 2012;63:3998. doi:10.3791/3998
39. Kozera B, Rapacz M. Reference genes in real-time PCR. J Appl Genet. 2013;54(4):391-406. doi:10.1007/s13353-013-0173-x
40. Wong G, Wong I, Chan K, Hsieh Y, Wong S. A Rapid and Low-Cost PCR Thermal Cycler for Low Resource Settings. PLoS One. 2015;10(7):e0131701. doi:10.1371/journal.pone.0131701
41. Malorny B, Hoorfar J, Bunge C, Helmuth R. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl Environ Microbiol. 2003;69(1):290-6. doi:10.1128/aem.69.1.290-296.2003
42. Gudnason H, Dufva M, Bang DD, Wolff A. Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007;35(19):e127. doi:10.1093/nar/gkm671
43. Bohaychuk VM, Gensler GE, McFall ME, King RK, Renter DG. A real-time PCR assay for the detection of Salmonella in a wide variety of food and food-animal matricest. J Food Prot. 2007;70(5):1080-7. doi:10.4315/0362-028x-70.5.1080
Authors
Copyright (c) 2021 Alfi Sophian, Ratna Purwaningsih, Muindar Muindar, Eka Putri Juniarti Igirisa, Muhammad Luthfi Amirullah

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Authors continue to retain the copyright to the article if the article is published in the Borneo Journal of Pharmacy. They will also retain the publishing rights to the article without any restrictions.
Authors who publish in this journal agree to the following terms:
- Any article on the copyright is retained by the author(s).
- The author grants the journal the right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share work with an acknowledgment of the work authors and initial publications in this journal.
- Authors can enter into separate, additional contractual arrangements for the non-exclusive distribution of published articles (e.g., post-institutional repository) or publish them in a book, with acknowledgment of their initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their websites) prior to and during the submission process. This can lead to productive exchanges and earlier and greater citations of published work.
- The article and any associated published material are distributed under the Creative Commons Attribution-ShareAlike 4.0 International License.